Step 1: Review the incoming data
The first step after your merge request is created is verifying the incoming data. Any issues with MRI data should be resolved as quickly as possible.
A lot of information is provided in the merge request interface, we will guide you through the information step by step, and then explain how specific issues can be dealt with. Always feel free to contact your data steward.
1. Check the data overview
The table at the top of the merge request provides basic information about the converted data, including the original tar file name, and subject information. Review this information.
2. Check the activity log
Scroll down in the the merge request, under the header activity there should be a series of messages that were generated during the conversion process. Carefully review the following:
- Is anything missing for the sorted DICOM files?
- Check the BIDScoin mapping log and BIDScoin conversion log for ERRORS
- Are volumes discarded correctly (6 volumes are discarded as a standard)?
3. Check BIDS validator output
To review the output of the BIDS validator, click the first circle on the right side of the pipeline overview:
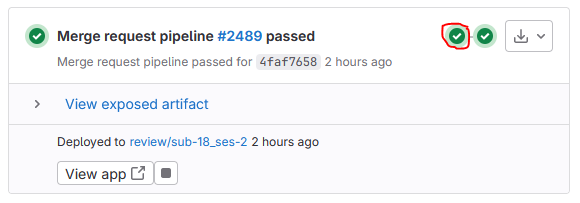
- Click bids-validate in the appearing box
You will see the output of the BIDS validator. Pay attention to the following:
- Does the validator report any errors?
- Missing event data is expected, we will add this later
- Make note of errors related to inconsistencies or corrupt data
If you need additional information you can also look at the full log file.
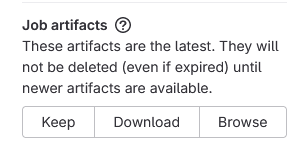
- Select Browse under job artifacts in the right hand panel
- Follow the link on the next page
- Download the
bidsvalidation.logfile
3. Handle conversion issues
Depending on the issue it might be possible to fix it in the merge request. It is possible the data needs to be exported again from the scanner, or in the worst case, if there are issues with the data itself, it must be collected again or discarded. If you have any questions you can always contact a dataset steward, leave a comment in a relevant issue or in the Dataset Status.
Missing or corrupted files
If any files are missing, or files are corrupted (the BIDS validator should raise issues concerning unreadable files, issues in nifti headers, etc.), contact your data steward. It is likely that files will need to be exported again from the scanner. We will assist you in this.
Invalid files or additional files
If files were exported that should not have ended up in the repository, they should be removed before merging your data. This can also happen when multiple versions of the same sequence were exported because of concerns about data quality. The assessment about which file to keep should be done on the merge request, and other files should be deleted before merging in. In most cases, it is not enough to simply delete the files. Thus, see the guide on how to delete invalid files.
Wrongly named files
If there are wrongly named files in the merge request, it might not be possible to fix it directly in the webIDE. If one of the files that should be renamed is a large file, such as a nifti file, it is necessary to clone your dataset. Files can then be renamed on your local system, and these changes should be pushed back to the repository. See our guide on working on data locally.
Inconsistent parameters
If sequences were collected with the wrong parameters, ensure they can still be used. Make note of the inconsistency in the README.md, specifically the missing data section. You can also comment the information about the inconsistent parameters directly in the "Fill in README.md" issue, if you do not plan to work on your README.md file until later. If data is not usable, this merge request can be deleted.